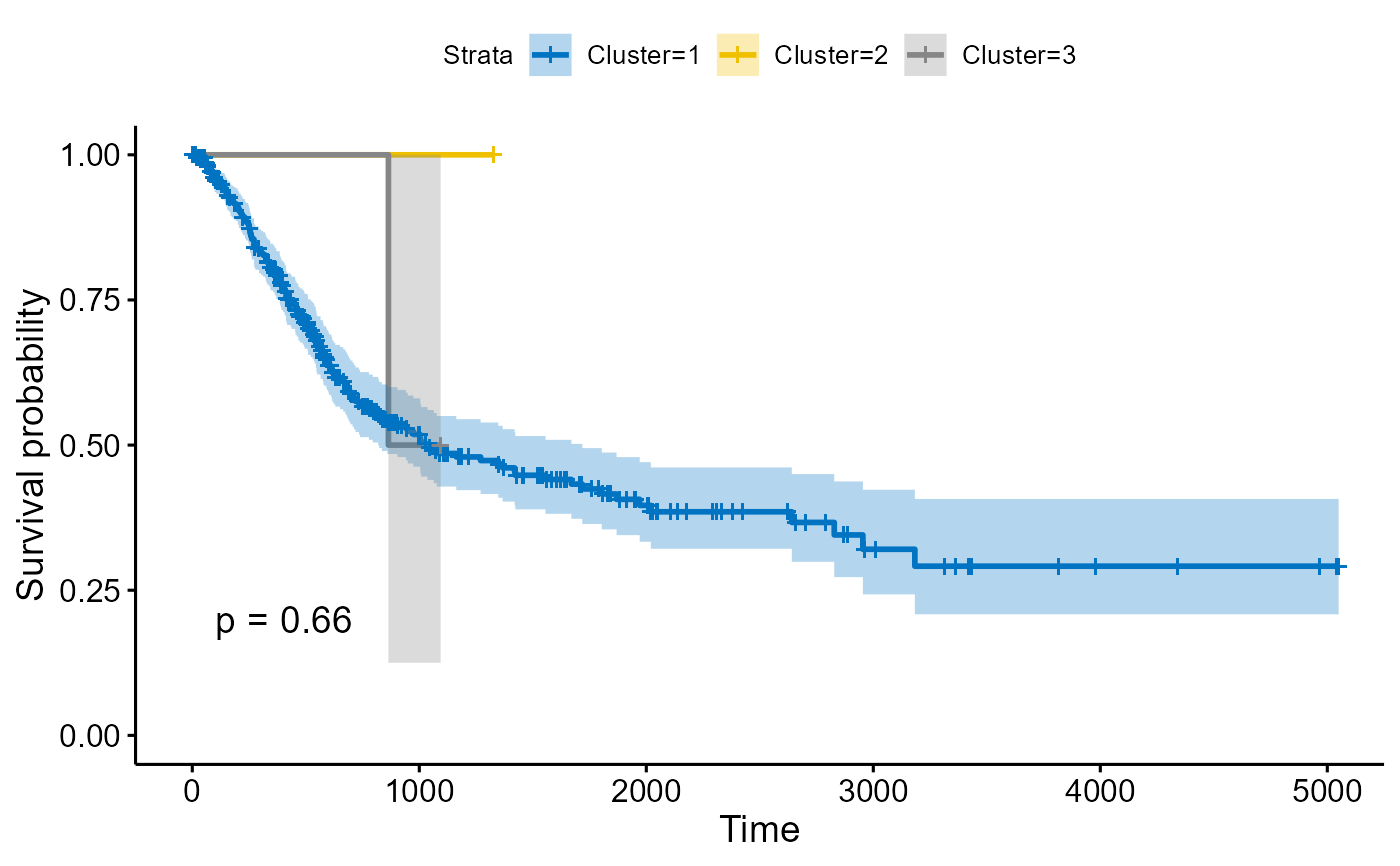
Surplot
Surplot.RdDraw a KM survival map
Arguments
- results
The results after clustering
- meta
The clinical data downloaded from TCGA
- optK
The optimal K
Examples
data(results)
data(meta)
maxK = 6
Kvec = 2:maxK
x1 = 0.1; x2 = 0.9 # threshold defining the intermediate sub-interval
PAC = rep(NA,length(Kvec))
names(PAC) = paste("K=",Kvec,sep="") # from 2 to maxK
for(i in Kvec){
M = results[[i]]$consensusMatrix
Fn = ecdf(M[lower.tri(M)])
PAC[i-1] = Fn(x2) - Fn(x1)
}
optK = Kvec[which.min(PAC)]
Surplot(results,meta,optK=3)